Create balanced clusters of equal size
balanced_clustering(x, K, method = "centroid", solver = NULL)Source
The centroid method was originally developed and contributed by Meik Michalke. It was later rewritten by Martin Papenberg, who also implemented the integer linear programming method.
Arguments
- x
The data input. Can be one of two structures: (1) A feature matrix where rows correspond to elements and columns correspond to variables (a single numeric variable can be passed as a vector). (2) An N x N matrix dissimilarity matrix; can be an object of class
dist(e.g., returned bydistoras.dist) or amatrixwhere the entries of the upper and lower triangular matrix represent pairwise dissimilarities.- K
How many clusters should be created.
- method
One of "centroid" or "ilp". See Details.
- solver
Optional. The solver used to obtain the optimal method if
method = "ilp". Currently supports "glpk" and "symphony". Is ignored formethod = "centroid".
Value
An integer vector representing the cluster affiliation of each data point
Details
This function partitions a set of elements into K
equal-sized clusters. The function offers two methods: a heuristic
and an exact method. The heuristic (method = "centroid")
first computes the centroid of all data points. If the input is a
feature matrix, the centroid is defined as the mean vector of all
columns. If the input is a dissimilarity matrix, the most central
element acts as the centroid; the most central element is defined
as the element having the minimum maximal distance to all other
elements. After identifying the centroid, the algorithm proceeds as
follows: The element having the highest distance from the centroid
is clustered with its (N/K) - 1 nearest neighbours
(neighbourhood is defined according to the Euclidean distance if
the data input is a feature matrix). From the remaining elements,
again the element farthest to the centroid is selected and
clustered with its (N/K) - 1 neighbours; the procedure is
repeated until all elements are part of a cluster.
An exact method (method = "ilp") can be used to solve
equal-sized weighted cluster editing optimally (implements the
integer linear program described in Papenberg and Klau, 2020;
(8) - (10), (12) - (13)). The cluster editing objective is the
sum of pairwise distances
within clusters; clustering is accomplished by minimizing this
objective. If the argument x is a features matrix, the
Euclidean distance is computed as the basic unit of the cluster
editing objective. If another distance measure is preferred, users
may pass a self-computed dissimiliarity matrix via the argument
x.
The optimal method = "ilp" uses a "solver" to optimize
the clustering objective. See optimal_anticlustering
for an overview of the solvers that are available.
References
Grötschel, M., & Wakabayashi, Y. (1989). A cutting plane algorithm for a clustering problem. Mathematical Programming, 45, 59–96.
Papenberg, M., & Klau, G. W. (2021). Using anticlustering to partition data sets into equivalent parts. Psychological Methods, 26(2), 161–174. https://doi.org/10.1037/met0000301.
Examples
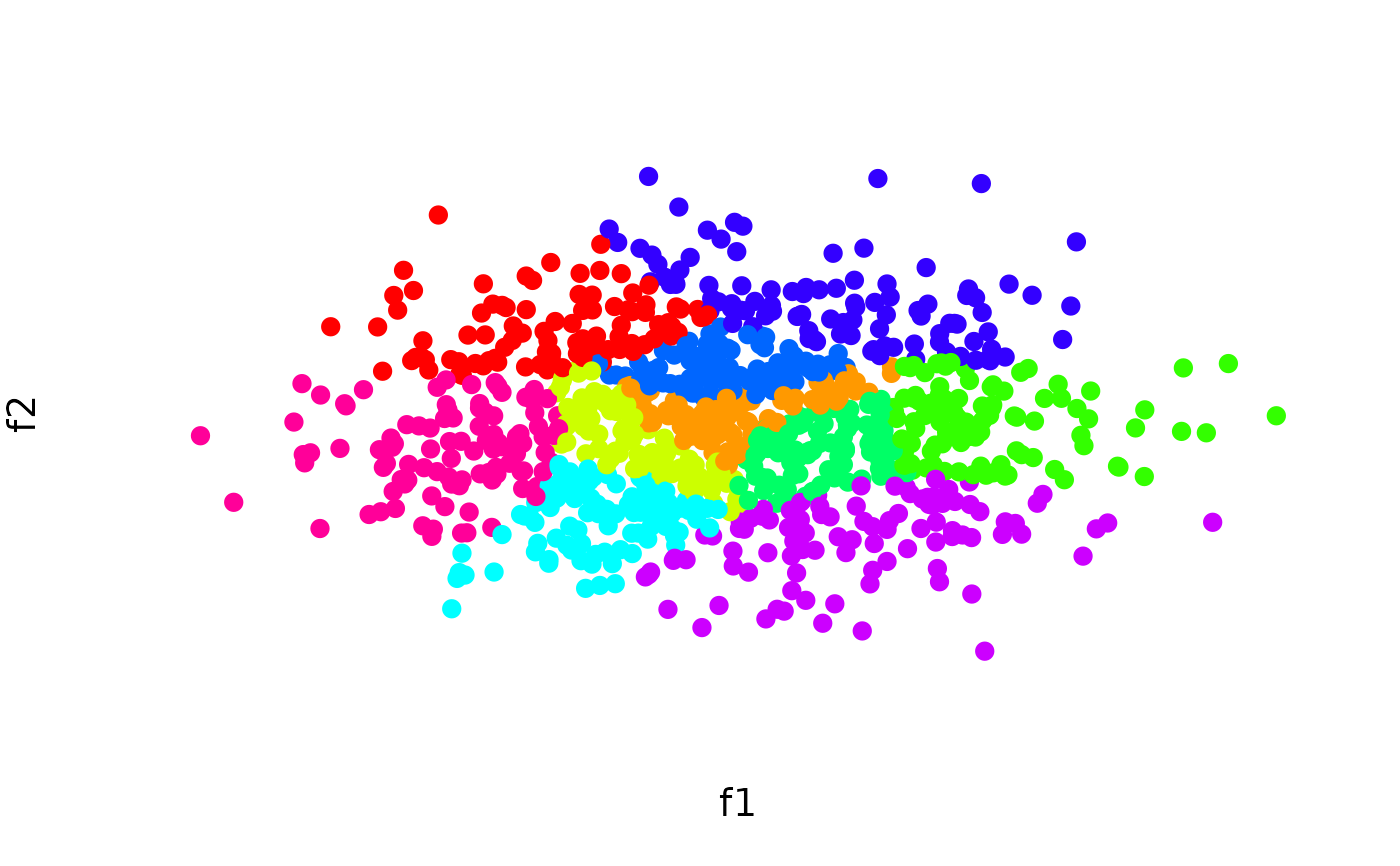
# Cluster a data set and visualize results
N <- 1000
lds <- data.frame(f1 = rnorm(N), f2 = rnorm(N))
cl <- balanced_clustering(lds, K = 10)
plot_clusters(lds, clusters = cl)
 # Repeat using a distance matrix as input
cl2 <- balanced_clustering(dist(lds), K = 10)
plot_clusters(lds, clusters = cl2)
# Repeat using a distance matrix as input
cl2 <- balanced_clustering(dist(lds), K = 10)
plot_clusters(lds, clusters = cl2)